Back to 2024 Abstracts
DISTINCT MATRISOME EXPRESSION PATTERNS IN DIFFERENT PDAC METASTATIC SITES
Lorena Llontop
*1, Maren Drenckhan
1, Thorben Sauer
2, Louisa Bolm
1, Rüdiger Braun
1, Olga Lapshyna
1, Meike ten Winkel
1, Steffen Deichmann
1, Axel Künstner
1, Hauke Busch
2, Ulrich F. Wellner
1, Susanne Sebens
3, Andrew S. Liss
4, Timo Gemoll
2, Tobias Keck
1, Kim C. Honselmann
11Department of Surgery, Lübeck, Schleswig-Holstein, Germany; 2Universitat zu Lubeck, Lubeck, Schleswig-Holstein, Germany; 3Christian-Albrechts-Universitat zu Kiel, Kiel, Schleswig-Holstein, Germany; 4Massachusetts General Hospital Department of Surgery, Boston, MA
Pancreatic ductal adenocarcinoma (PDAC) is a leading cause of cancer deaths with a survival rate of less than 11%. A prominent feature of PDAC is its highly desmoplastic reaction, characterized by an abundance of extracellular matrix (ECM) in primary and metastatic tumors. This study focuses on analyzing the PDAC tumor microenvironment in primary and metastatic tumors.
Statistical analysis of publicly available RNA expression profiles of PDAC primary (n = 145) and metastatic (liver [n = 25], lung [n = 8], lymph nodes [n = 9], and peritoneal [n = 7]) tumors was performed. In addition, primary (n = 5) and liver metastatic (n = 6) PDAC patient-derived xenograft (PDX) mouse models were analyzed using paired-end NextGen sequencing. Species-specific differences facilitated the identification of genes expressed and altered in both the stroma (mouse) and cancer cells (human).
Gene set enrichment analysis revealed multiple ECM-related gene sets significantly regulated between primary and metastatic sites in liver, lung, and lymph nodes, and peritoneal metastases (Fig. 1). A similar number of ECM genes were expressed in primary tumors (n = 416), peritoneum (n = 377), lymph nodes (n = 363), liver (n = 353), and lung (n = 350) metastases, with significant overlap in expression between all five sites (317 common genes). However, ECM genes exhibited substantial differential expression (-logFC > 1.3 or <1.3, q<0.05) between primary and metastatic sites. Moreover, the extent of differential expression correlated with the distance from the primary site, with peritoneal metastasis showing a 6% alteration, lymph nodes 13%, liver 35%, and lung 43% (Fig. 2). These findings were partly confirmed by PDX analysis, which displayed significant differential expression of ECM genes between primary and liver metastasis, encompassing 58% of all expressed ECM genes. Interestingly, within the stromal compartment, most genes were down-regulated in metastasis (57% down, 37% up), while in cancer cells, most genes were up-regulated (61% up, 23% down), suggesting distinct changes in ECM components in PDAC metastasis.
Altogether, exploratory analysis of ECM gene expression in PDAC metastases revealed unique alterations in ECM components between primary and metastatic tumors. These findings suggest that PDAC ECM changes are specific to each metastatic tumor site and cell origin, offering a multitude of potential therapeutic targets for both the stromal and cancer-specific components in PDAC metastasis.
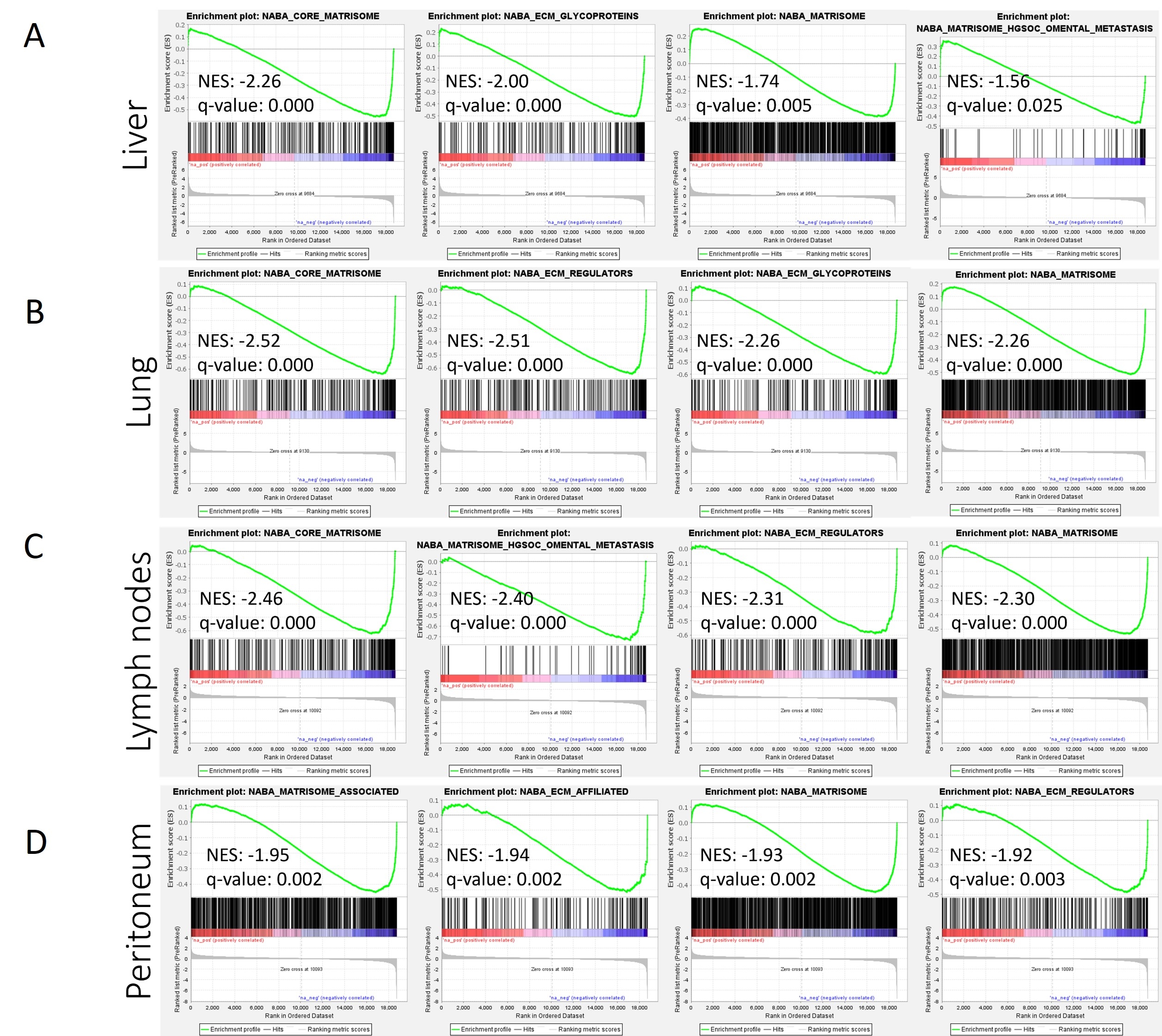 Figure 1. GSEA analysis between primary and metastatic PDAC tumors.
Figure 1. GSEA analysis between primary and metastatic PDAC tumors. C2 (curated gene sets) reference gene set was on all genes.
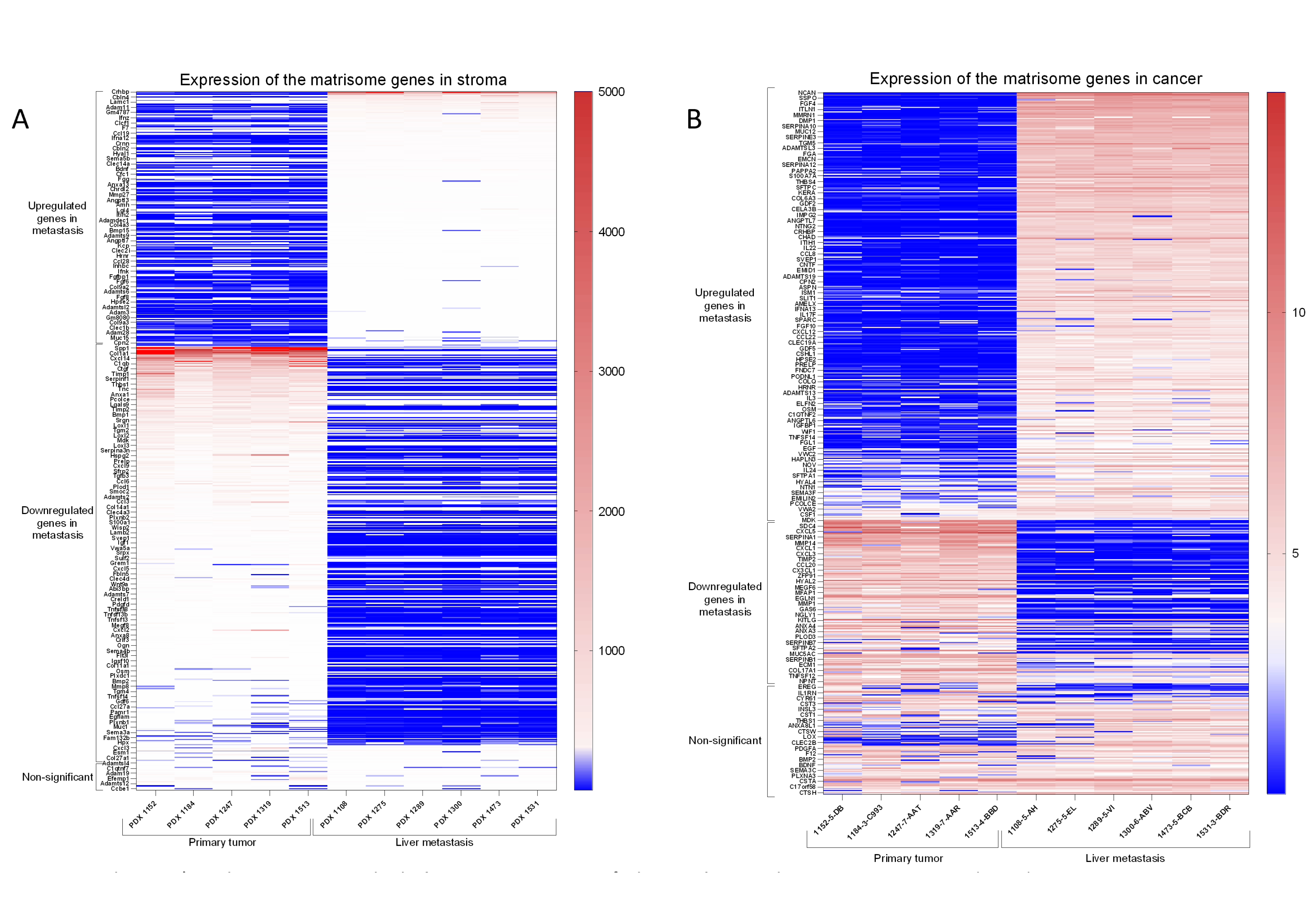 Figure 2. A) Matrisome genes expression in the stroma compartment of primary and metastatic PDAC PDX tumors.
Figure 2. A) Matrisome genes expression in the stroma compartment of primary and metastatic PDAC PDX tumors. From the total 1105 mouse matrisome genes, 682 were detected in both, primary tumor and liver metastasis. From these, 236 upregulated genes were found in metastasis, 383 downregulated in metastasis and 63 had non-significant variation in expression between primary and metastasis.
B) Matrisome genes expression in the tumor of primary and metastatic PDAC PDX tumors. From the total 1062 human matrisome gene list, 633 were detected in both, primary tumor and liver metastasis. From these, 383 upregulated genes were found in metastasis, 149 downregulated in metastasis and 101 had non-significant variation in expression between primary and metastasis.
Back to 2024 Abstracts

