STOOL DNA-BASED SYNDECAN-2(SDC2) METHYLATION TESTING FOR THE EARLY DETECTION OF COLORECTAL CANCER IN AN AVERAGE RISK POPULATION
Suk-Hwan Lee*1, Youn Young Park1, Chang Woo Kim2, Hyunjin Kim3, Hyoung Rae Kim4, Daeyoun D. Won5, Woojung Nam5, Hyung Jin Kim6, Bong-Hyeon Kye8, Sungwhan An7, Tae-Jeong Oh7
1Surgery, Kyung Hee University Hospital at Gangdong, Gangdong-gu, Seoul, Korea (the Republic of); 2Ajou University School of Medicine and Graduate School of Medicine, Suwon, Gyeonggi-do, Korea (the Republic of); 3Koo Hospital, Daegu, Korea (the Republic of); 4Busan Hangun Hospital, Busan, Korea (the Republic of); 5Seoul Songdo Hospital, Seoul, Seoul, Korea (the Republic of); 6Catholic University of Korea Eunpyeong St Mary's Hospital, Seoul, Seoul, Korea (the Republic of); 7Genomictree, Daejeon, Korea (the Republic of); 8Catholic University of Korea Suwon St. Vincent's Hospital, Suwon, Korea (the Republic of)
Background and Aims
A cost-effective, accurate and sensitive non-invasive detection tool for colorectal cancer (CRC) is urgently needed to reduce disease-related mortality and incident rate. SDC2 methylation frequently occurs in all stages of CRC. This study aims to evaluate a stool-based SDC2 DNA methylation test to detect CRC in a prospective clinical study. This study is registered in clinicaltrial.gov (NCT04304131)
Methods
A prospective multicenter study was designed to evaluate the clinical performance of the test measuring the methylation status of SDC2 DNA in stool by real-time PCR employing Linear Target Enrichment and Quantitative Methylation-Specific PCR (LTE-qMSP SDC2). We used a pre-specified cutoff value based on a training study to determine SDC2 methylation positivity in stool samples. Stool samples were collected from an average risk population of asymptomatic individuals ? 60 years prior to colonoscopy. The testing results were independently analyzed by comparison with colonoscopic findings and pathology outcomes as reference standards. The primary outcome measures for detecting CRC were sensitivity and specificity. For PPV and NPV calculation, the prevalence of CRC was assumed to be 2.0% based on this work.
Results
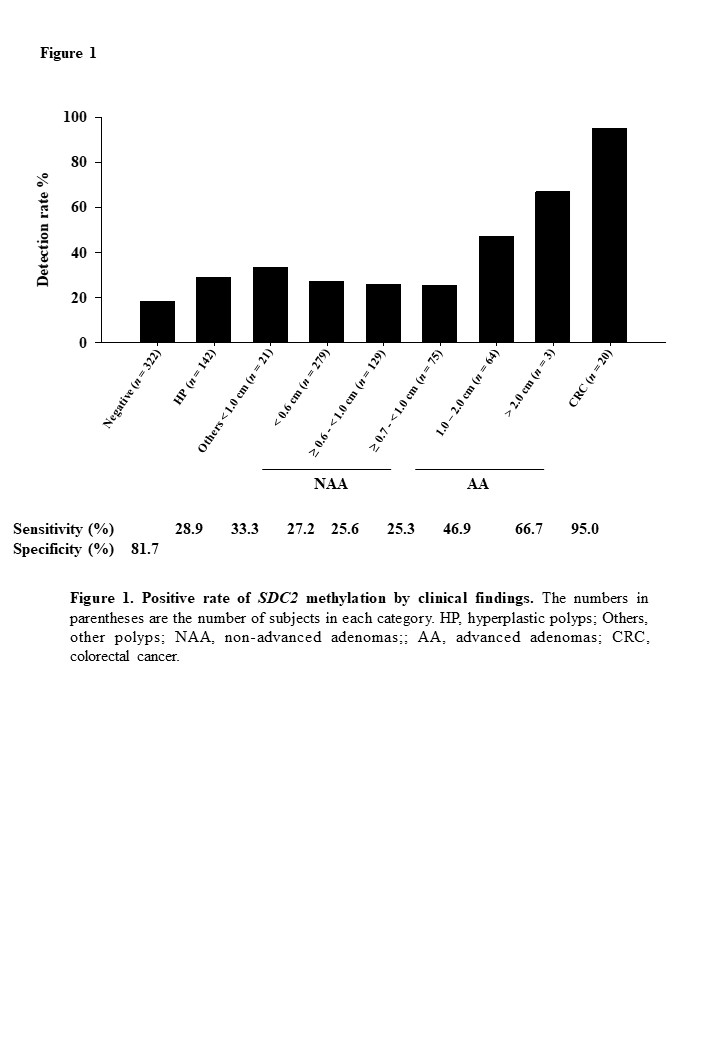
Of the 980 evaluable participants, had 20 (2.0%) CRC, 67 (6.8%) had advanced adenomatous polyps (AA, ?1.0 cm), and 571 (58.3%) had non-advanced polyps (NAP, <1.0 cm), and 322 patients (32.9%) were negative on colonoscopy examination. The sensitivity of LTE-qMSP SDC2 test for CRC and AA was 95% (19/20) (95% CI 75.1-99.9%) and 47.8% (32/67) (95% CI 35.4-60.4%), respectively. The specificity is 81.7% (59/322) (95% CI 77.0-85.8%) based on the finding on a colonoscopy (Figure 1). The sensitivity for early stages (0-II) of CRC was 92.3% (12/13). The sensitivity for AA and CRC was 58.6% (51/87, 95% CI 47.5-69.1%). The sensitivities for ?0.6 cm adenomas and CRC, and ?0.7 cm adenomas and CRC were 38.9% (84/216, 95% CI 32.4-45.7%) and 43.2% (70/162, 35.5-51.2%), respectively (Table 1). PPV and NPV were 9.6% (95% CI 7.6-12.0%) and 99.9% (95% CI 99.2-100%), respectively. The sensitivity of the test for adenomas increased as the lesion size increased and was not associated with CRC stage (P = 0.566).
Conclusion
Non-invasive SDC2 methylation test in stool DNA by LTE-qMSP attained a high sensitivity for CRC detection in an average-risk population.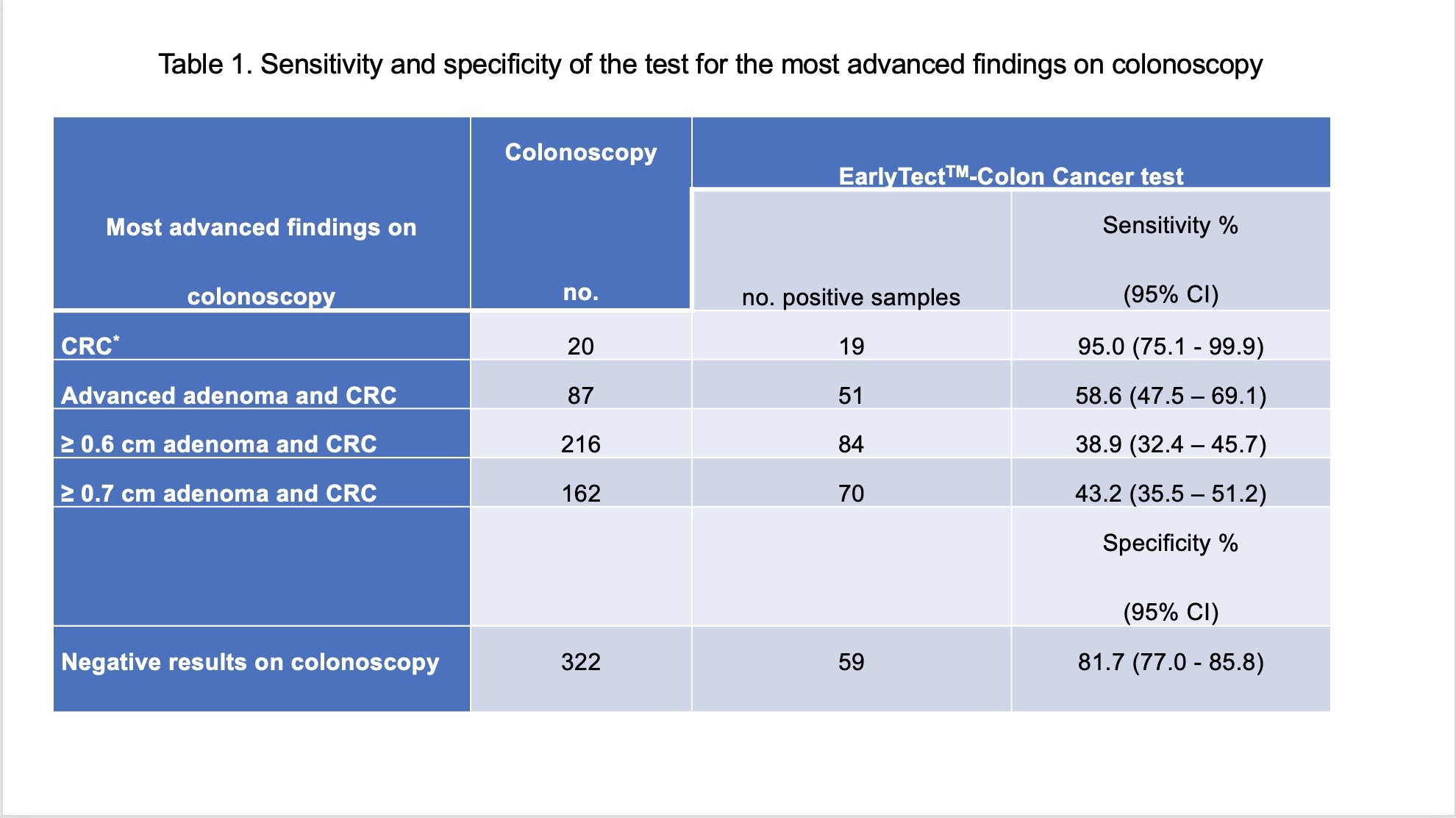
Back to 2022 Abstracts
