|
Back to 2017 Program and Abstracts
MODIFICATION OF THE APPENDIX MICROBIOME IN ACUTE APPENDICITIS AND COLON ADENOCARCINOMA
Alasdair Scott*, James L. Alexander, Julian Marchesi, James M. Kinross
Surgery and Cancer, Imperial College London, London, United Kingdom
Background
The appendix had been thought to be a vestigial organ, devoid of function. However, mounting evidence suggests that the appendix plays a role in maintaining gastrointestinal microbial ecology and preventing dysbiosis by acting as a “reservoir” for beneficial bacteria. We therefore aimed to compare the healthy appendix microbiome with the pathological microbiome associated with acute appendicitis and colonic adenocarcinoma.
Methodology
Adult patients undergoing appendicectomy for suspected appendicitis or ileocaecal resection for right-sided adenocarcinoma at a London teaching hospital were prospectively recruited. Tissue was sampled from the appendix in all patients and additionally from the tumour and adjacent healthy mucosa in patients with adenocarcinoma. DNA was extracted from samples and V1-2 regions of the 16S rRNA gene were sequenced on the Illumina MiSeq platform. Data were analysed by Mothur to determine OTU abundance per sample. Species assignment was performed using NCBI BLAST. Multivariate models were constructed in SIMCA to identify key differentiating species. Univariate comparison of the relative abundance of identified species was performed in STAMP. Final diagnosis of appendicitis was based on histological evidence of transmural neutrophilic infiltration.
Results
54 patients were included: 37 with suspected appendicitis and 17 with colon adenocarcinoma. Of the patients with suspected appendicitis, 23 (62%) patients had histological evidence of appendicitis (with 7 perforation) while 14 (38%) did not. Supervised multivariate analysis could differentiate healthy and inflamed appendixes (Figure 1, OPLS-DA, R2=0.62, Q2=0.17). Compared to healthy appendixes, inflamed appendix tissue had reduced abundance of gut commensals including Coprococcus species (p<0.001), Ruminococcus lactaris (p=0.027) and Bifidobacterium longum (p=0.012) and was characterised by an overabundance of pathogenic bacteria typically found in the mouth including Campylobacter gracilis (p=0.019), Parvimonas micra (p=0.054), Prevotella tannerae (p=0.051) and Fusobacterium nucleatum (p=0.29). In contrast the, appendixes of patients with colon adenocarcinoma demonstrated an abundance of Clostridia species (p<0.02), Faecalibacterium prausnitzii (p=0.067) and Stenotrophomonas maltophilia (p=0.011). Interestingly, F. nucleatum, which has been associated with colorectal cancer, was found in higher abundance in the appendixes of patients with colorectal cancer than in healthy mucosa adjacent to the tumour (p=0.059).
Conclusion
Both acute appendicitis and colorectal carcinoma are associated with changes in the appendix microbiome. The reason for the preponderance of oral pathogens in acute appendicitis is unclear. The abundance of F. nucleatum in the appendixes of patients with colorectal cancer may reflect overexpression of this bacterium on the tumour.
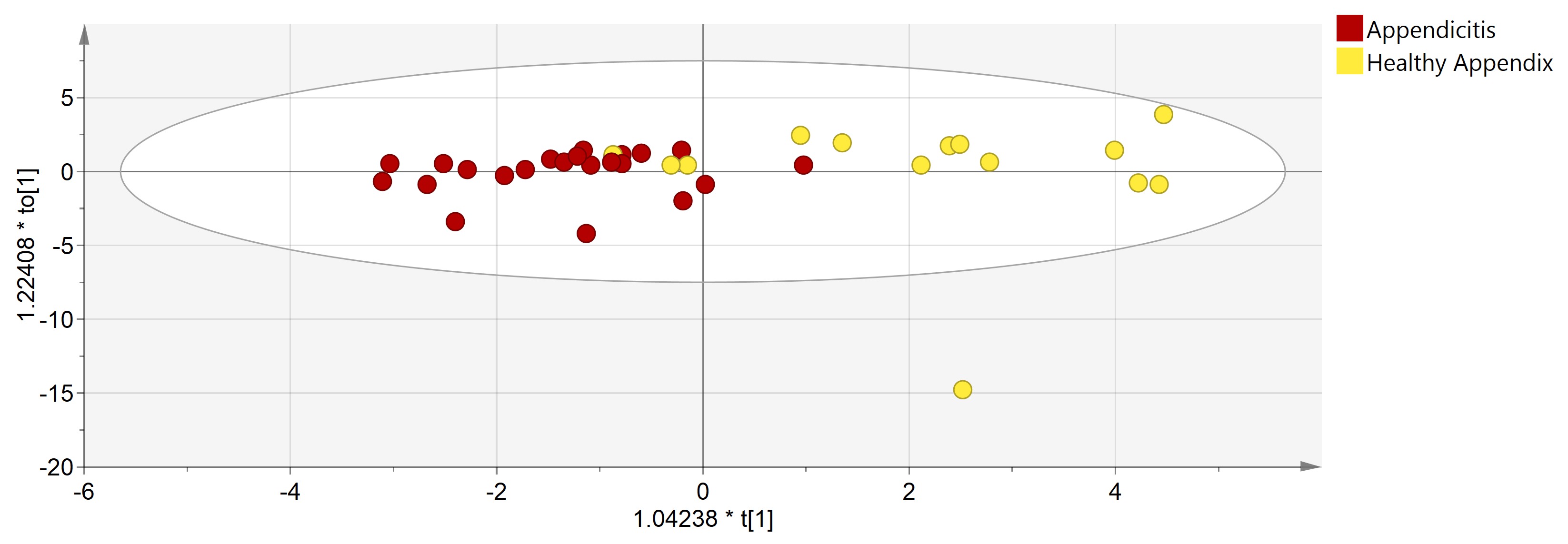
Scores plot of OPLS-DA model comparing microbiomes of normal (n=14) vs. inflamed appendixes (n=23). R2=0.62, Q2=0.17.
Back to 2017 Program and Abstracts
|


